In 2011, the National Academies of Sciences, Engineering, and Medicine released a major report titled, Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease [1]. Looking to develop a framework for classifying human disease based on molecular biology and epidemiology, the report laid out a roadmap for what would become known as precision medicine. The National Academies proposed that the U.S. government sequence the genomes of a million participants in order to begin developing ways of linking individual medical histories with broader insights drawn from genomic data. In 2015, the Departments of Defense, Health and Human Services, and Veterans Affairs acted upon this recommendation, initiating the “All of Us” million-genome sequencing effort.
Genomic data can aid the Department of Defense (DoD) and the warfighter in a multitude of ways. For example, genetic testing could allow military health personnel to precisely tailor dosages to the rate at which individual warfighters metabolize certain medications, allowing for precise pain management that reduces the potential for dependencies. Genomic data can also provide medical staff with key insights into mental health treatment. A 2018 study of the pharmacogenomics of 1,871 moderately to severely depressed patients found that genetic testing, when combined with targeted prescription, led to substantial improvements in depression outcomes [2]. As both of these examples illustrate, genomic data has the potential to significantly improve warfighter medical treatments and health across a range of medical needs.
Since the start of the All of Us project, hundreds of thousands of human genomes have been fully sequenced. Since the start of the All of Us project, hundreds of thousands of human genomes have been fully sequenced. In January 2017, Illumina—the largest sequencing firm—reported that 500,000 human genomes have been sequenced worldwide, up from just 65,000 two years before [3, 4]. Other groups, including the Eli & Edyth Broad Institute of the Massachusetts Institute of Technology and Harvard, and BGI (formerly the Beijing Genomics Institute), are also engaged in large-scale sequencing [5–7]. However, reductions in sequencing costs have presented new challenges related to high-volume data storage, distribution, and analysis [8]. Conservative estimates predict that 1 zettabyte of genomic sequencing data will be generated annually by 2025, requiring future data storage capabilities on the exabyte scale [8].
This volume of data may soon overload the ability of conventional on-site storage to hold and process genomic data. Because of this, a large amount of the current and future stored and processed genomic data is being handled by cloud service providers [9]. Genomic data at all levels of processing are potentially sensitive—capable of revealing characteristics of an individual’s identity, including information about sex, disease risk factors, race, and ethnic background. Tools and methods for improving the protection of genomic data are an important consideration for DoD and a requirement for secure operations.
Key Considerations
DoD is the first and most critical layer of homeland defense and security. As such, DoD must protect its warfighters and their associated data, as this work is directly related to national security. Therefore, DoD should be on the cutting edge of cybersecurity research and innovation, ensuring the protection of all systems on which warfighter data is transferred and located.
One major issue with securing genomic data is that its underlying digital infrastructure was built in an academic and generally open-access environment. This means that many of the standard protocols, software, and best practices need to account for risk and misuse. Protecting genomic data requires attention to several key issues:
(a) Securing the equipment: Including the sequencers, validation equipment, storage devices, etc.
(b) Assuming that all data providing genomic information (no matter how fragmented) contains information that can be used to target individuals.
(c) Maintaining data integrity over the life-cycle of the data—from creation, processing, analysis, and long-term storage.
(d) Assuming that software pipelines (which may contain a couple or dozens of individual programs) have not had a formal security audit, and that commercial solution packages use open source software.
(e) Understanding that data created or processed for other purposes (e.g., previous medical treatment) may have been handled on cloud servers (including foreign servers).
(f) Appreciating the dual-nature of precision treatment. (Precision treatment is a way of targeting interventions to an individual’s personal biochemistry, but this level of data should be protected to avoid data misuse.)
Any full-scale application of genomics in a security setting requires attention to these six issues. There is a clear need to protect genomic data through robust systems and measures. The rapid development of genomic data technology and the lack of full-scale security assessment in genomics has meant that many systems that handle critically important data possess under-studied risks. Furthermore, the piecemeal auditing of systemic parts may lead to gaps in security or reduced capabilities. Modeling and simulating the ways in which genomic data are stored, accessed, and retrieved for analysis is a useful method for testing genomic data systems.
Modeling and Assessment of Genomic Data
One approach to modeling and assessment is to develop a full-scale realistic model, sometimes called “genomics-in-a-box.” These models combine a realistic data source (i.e., human genomic analysis run through an analogous sequencing source), realistic throughput (hundreds to thousands of individuals at a time), and use of the same software and hardware components employed by genomic data systems. Human operators can function in the loop during these assessments, making decisions based on realistic scenarios. Researchers at Sandia National Laboratories have termed this modeling strategy Emulytics™.
Emulytics™ comes from the combination of “emulative network computing” and “analytics.” Emulytics™ provides the capability to combine real, emulated, and simulated devices into a single system-level experiment to answer a variety of cyber-related questions. Using Emulytics™, one can employ mechanisms to rapidly specify and deploy complex networked information systems of routers, switches, hosts, services, and applications, and integrate systems that can be configured and used for controlled experimentation as well as interactive exploration of system behavior [10].
The Emulytics™ testing approach has several notable features. First, it allows thorough red-teaming without the risk of damage to the primary systems. Second, Emulytics™ allows probing of software and hardware components as well as their interfaces (i.e., the critical and often fragile points where various components must interact). The Emulytics™ testing approach also provides for the assessment and wargaming of data exfiltration and manipulation. This process allows red-teams to “think like the enemy” and determine where the risks lay in infrastructure. The real challenge in this analysis is scaling to the immense amount of genomic data.
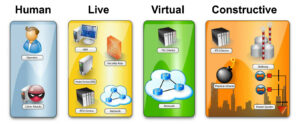
Figure 1. What is EmulyticsTM? Courtesy of Sandia National Laboratories.
Emulytics™ and Genomic Testing
One application of Emulytics™ in genomic security is through the modeling and assessment of large-scale genomic systems. These systems frequently require large suites of software and the interaction of multiple hardware components, handling terabytes of data per day. Incremental assessment may give a sense of which software are vulnerable, but not how these vulnerabilities affect the larger system.
Data Production and Hardware Modeling
A central aspect of Emulytics™ is data production and transfer. There are two main strategies for this production: (a) generation of realistic random data, and (b) the cloning of real data. There are a variety of methods for randomly generating realistic genomic data. Most of these rely on the random production of non-proprietary data formats (e.g., FASTQ, FASTA, SAM, VCF). The production of this data typically takes an initial data source (e.g., the reference human genome) and splits and randomizes the data, rearranging it into the desired format. The second approach involves mirroring traffic between hardware and software. This requires direct access to the raw data. This may be harder to collect in many instances, but has the distinct advantage of directly modeling the raw data producing equipment (e.g., an Illumina sequencer).
Software Library
A secondary aspect of designing and applying security tests, experiments, and audits is having a library of genomic software. There are no canonical genomic software workflows. Typically, workflows are built based on install suitability, experience with the software, available technical support, speed, performance, and accuracy. There are regularly hundreds of distinct software options strung across each of dozens of steps. Determining the risks posed by a particular software choice oftentimes means having not only the software, but also the version in place. Emulytics™ allows efficient pipeline creation, which can then allow plug-and-play pull and switch for different software alternatives. This is primarily done because there is an existing genomic software library.
External Resources
Oftentimes external resources are included in various aspects of a genomics pipeline. From the systems perspective, a cloud server is simply a storage and processing resource that communicates across particular protocols and limits certain aspects of resource function. Emulytics™ allows quick creation of external resources. These can be partitioned to investigate different security scenarios. For instance, many of the reference genomes are stored on FTP servers, without encryption. This may create security problems that can be investigated during assessment. Analysts can also, for example, investigate the security vs. performance trade-offs that occur when offboarding fundamental operations to cloud service providers.
Vulnerability Replay
One means of experimental assessment of vulnerabilities is to follow a hypothesis-guided approach. This is the model that Emulytics™ employs. To determine the impact of a known vulnerability, it is important to know what can be held constant, and which vulnerabilities change under evolving conditions. There are issues of scale that create new vulnerabilities, usually owing to automation and decreased visibility. There are also vulnerabilities that may affect entire classes of operations—for example, a flawed data format that can be identified from testing and assessing a variety of software options. Directing testable security hypotheses requires an ability to make assumptions about which part of the system can be held constant and to create and test alternative scenarios by varying other parts of the system.
Conclusion
The DoD can utilize genomic testing for personalized health management, particularly for military forces. For example, there is great potential in using genomic data to manage pain and mental health. However, keeping access to genomic data secure and safe requires DoD-led research and development.
Protocols, processes, and security plans must be established in order to provide access to genomic data by authorized parties and limiting access only to those individuals. Emulytics™ is one methodology that can be leveraged to expand genomic security research and development by understanding where the vulnerabilities are at different levels of scale, and building solutions and mitigation plans to address them. The overall goal for genomic security is for warfighters to keep their data secure but accessible to those who are assisting the mission. Cybersecurity experimentation through Emulytics™ could help to continuously move us in that direction.
Disclaimer
Supported by the Laboratory Directed Research and Development program at Sandia National Laboratories, a multi-mission laboratory managed and operated by National Technology and Engineering Solutions of Sandia, LLC, a wholly owned subsidiary of Honeywell International, Inc., for the U.S. Department of Energy’s National Nuclear Security Administration under contract DE-NA0003525. This paper describes objective technical results and analysis. Any subjective views or opinions that might be expressed in the paper do not necessarily represent the views of the U.S. Department of Energy or the United States Government. SAND2018-13633 J.
References
1. National Research Council (2011). Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, D.C., The National Academies Press. doi: 10.17226/13284
2. Tanner, J.-A., et al. (2018). Combinatorial pharmacogenomics and improved patient outcomes in depression: Treatment by primary care physicians or psychiatrists. Journal of psychiatric research 104: 157-162. doi: 10.1016/j.jpsychires.2018.07.012
3. Herper, M. (2017). Illumina Promises to Sequence Human Genome for $100 – But Not Quite Yet. Forbes. Retrieved from https://www.forbes.com/sites/matthewherper/2017/01/09/illuminapromises-to-sequence-human-genome-for-100-but-not-quite-yet/#6971dac8386d
4. Gebelhoff, R. (2015). Sequencing the genome creates so much data we don’t know what to do with it. The Washington Post. Retrieved from https://www.washingtonpost.
com/news/speaking-of-science/wp/2015/07/07/sequencing-the-genomecreates-so-much-data-we-dont-know-whatto-do-with-it/?utm_term=.494c19cdf76d
5. Broad Communications (2018). Broad Institute Sequences its 100,000th whole human genome on National DNA Day. Broad Communications. Retrieved from https://www. broadinstitute.org/news/broad-institute-sequences-its-100000th-whole-human-genome-national-dna-day
6. BGI (2018). “Genome DECODE Program.” Genome DECODE Program. Retrieved 20 August 2018, from https://gdp.bgi.com/page/index/en/.
7. Helmy, M., et al. (2016). Limited resources of genome sequencing in developing countries: Challenges and solutions. Applied and Translational Genomics 9: 15-19. doi:10.1016/j.atg.2016.03.003
8. Stephens, Z. D., et al. (2015). Big data: astronomical or genomical? PLoS biology 13(7): e1002195. doi: 10.1371/journal.pbio.1002195
9. Langmead, B. and A. Nellore (2018). Cloud computing for genomic data analysis and collaboration. Nature Review Genetics 19:208-219.
10. Urias, V., et al. (2015). Emulytics™ at Sandia National Laboratories. MODSIM World, Virginia Beach, VA.


